NMR
For NMR analysis, we currently support:
- when using ChemSpectra: JCAMP-DX files (*.jcamp, *.dx, *.jdx), zip (Bruker FID and BagIt) files.
- when using NMRium: JCAMP-DX files (*.jcamp, *.dx, *.jdx), zip (Bruker FID), Jeol (*.jdf), NMRium (*.nmrium) files.
When an NMRium analysis is saved, a .nmrium file and a preview image (SVG) are stored in Chemotion ELN.
The .nmrium file contains references to the associated spectra and the analysis modifications. The raw spectral data remains stored in the original .jdx or .zip files.
When ChemSpectra is available, an additional '*.edit.jdx' file may be stored for 1D NMR data. This enables the analysed data to be opened and continued in both ChemSpectra and NMRium.
Chemotion ELN supports data synchronisation between ChemSpectra and NMRium when both services are available.
Analysis using ChemSpectra
We currently support 1H, 13C, 15N, 19F, 29Si, and 31P NMR techniques.
-
To upload and view your data, you can follow this instruction.
-
Using tools bar to zoom in/out, add peaks, add multiplicity, etc.:
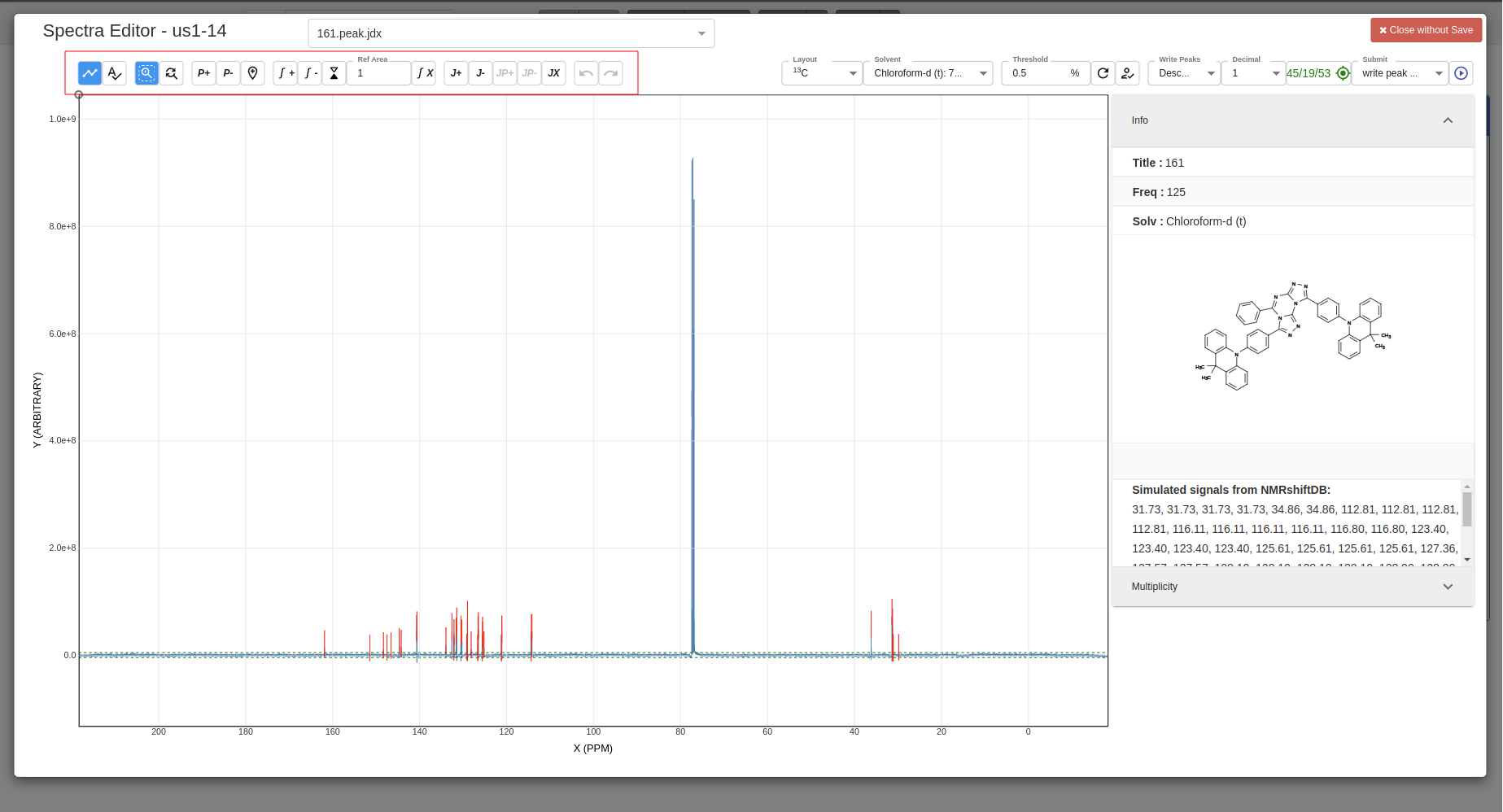
-
Example of added multiplicity:
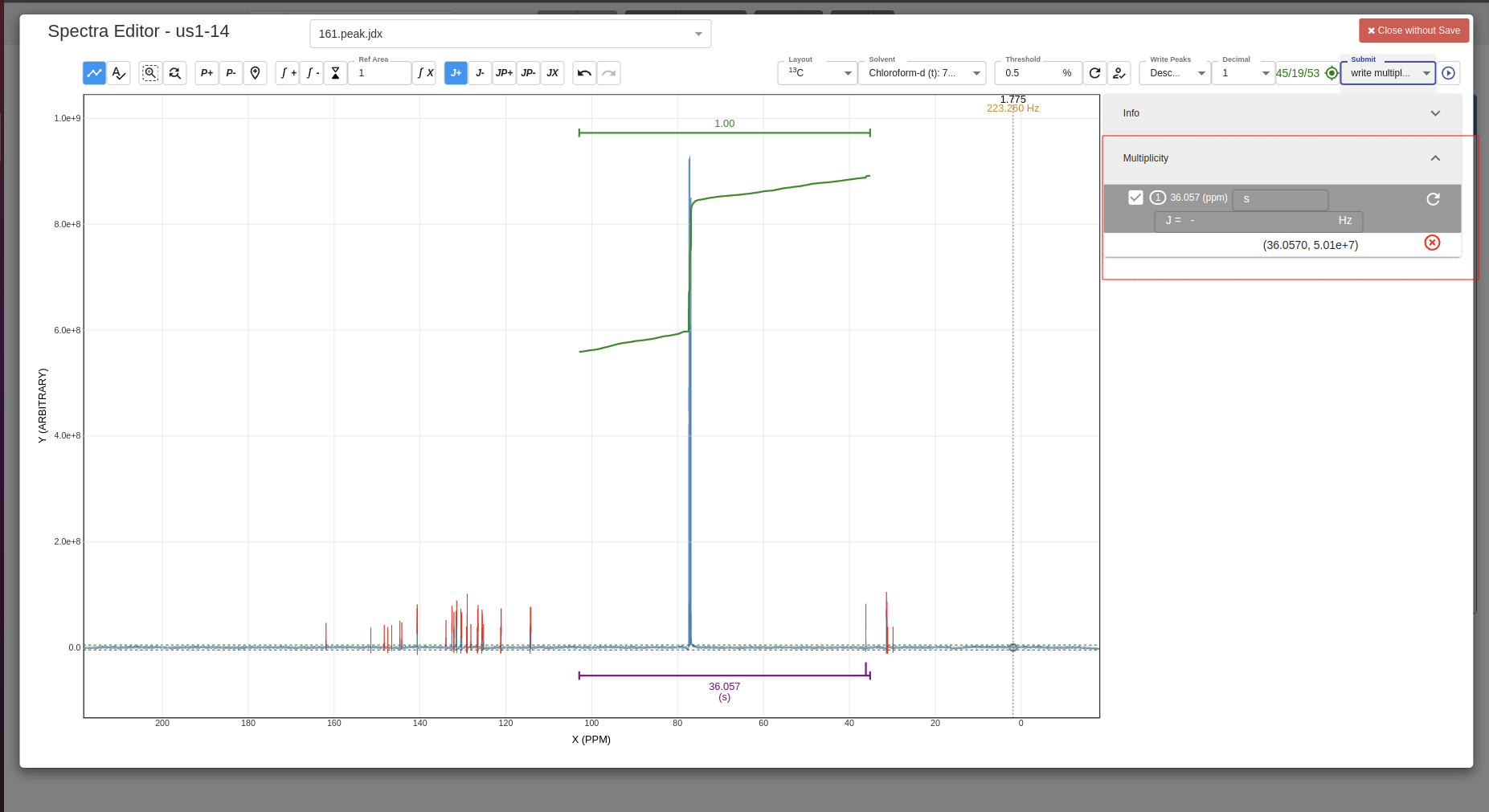
-
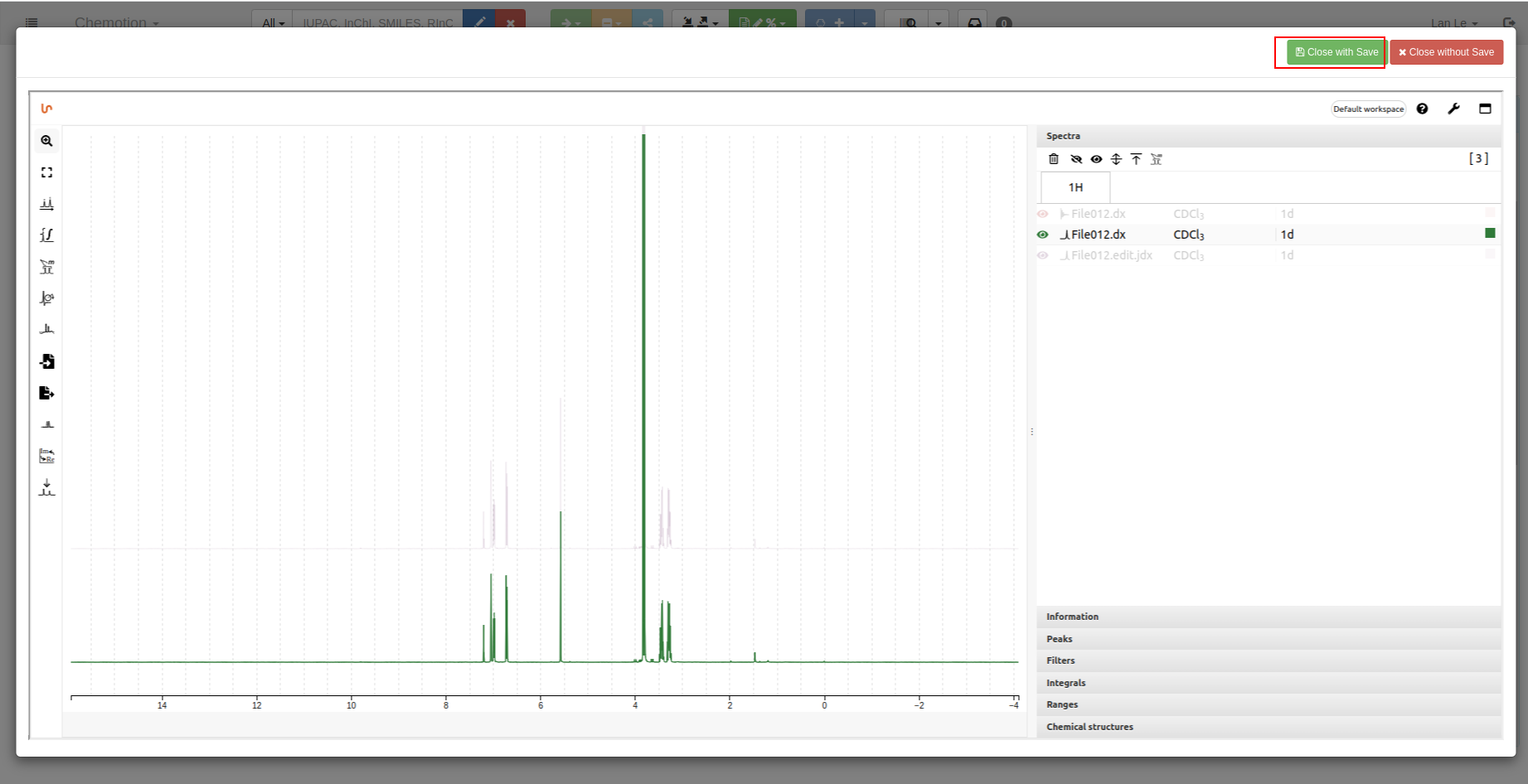
Select which data you want to be saved:
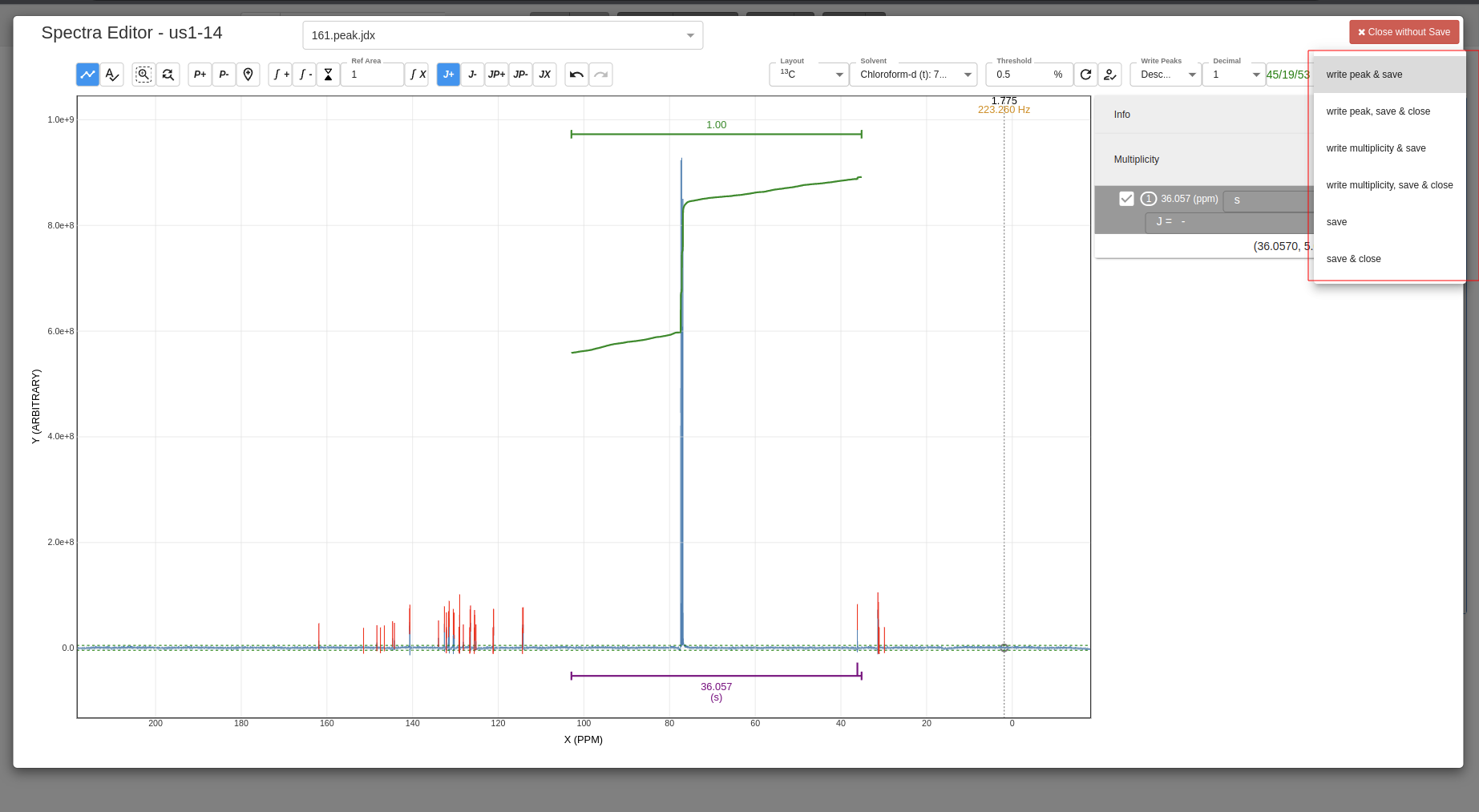
-
Confirm save:
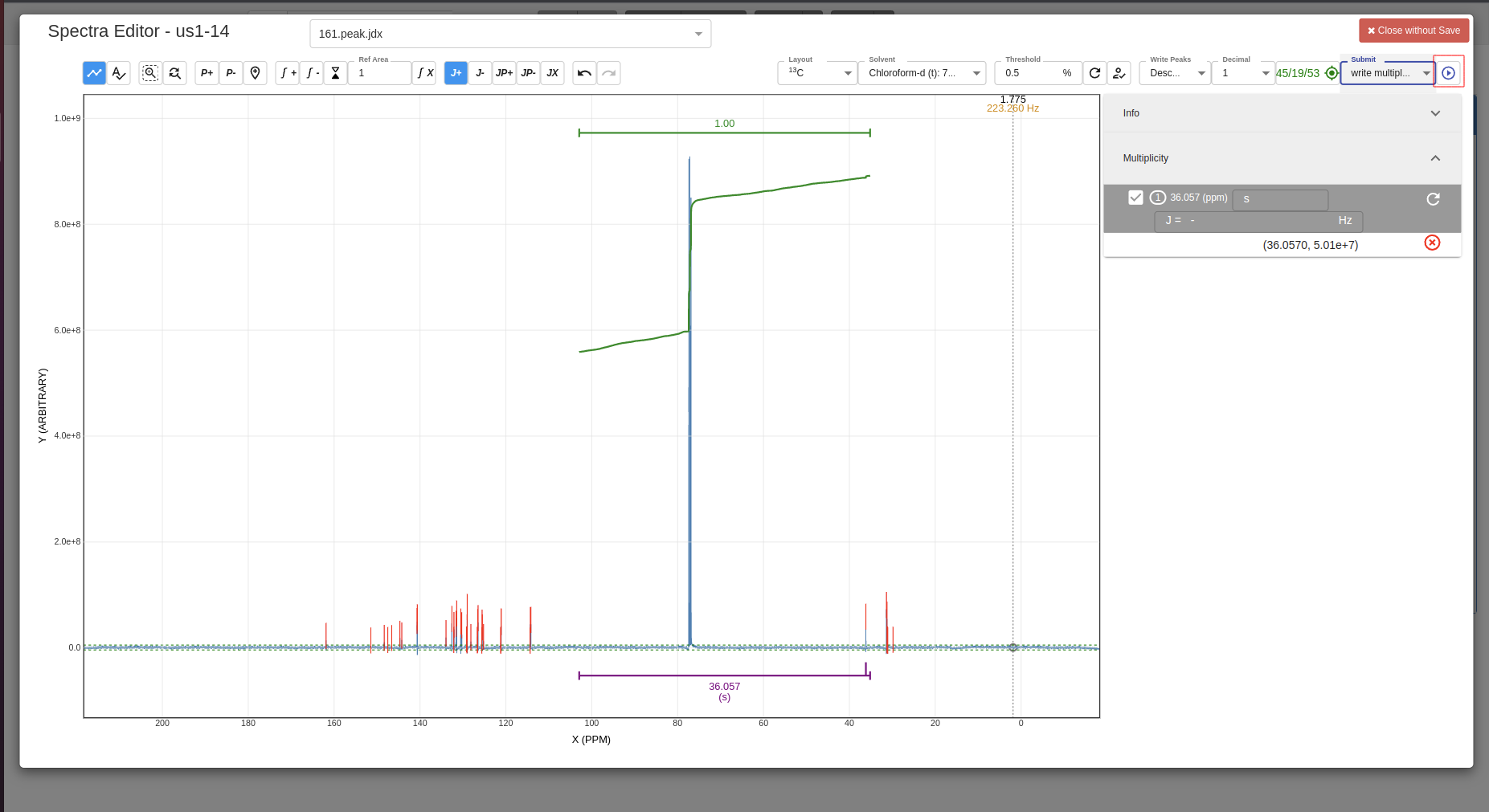
-
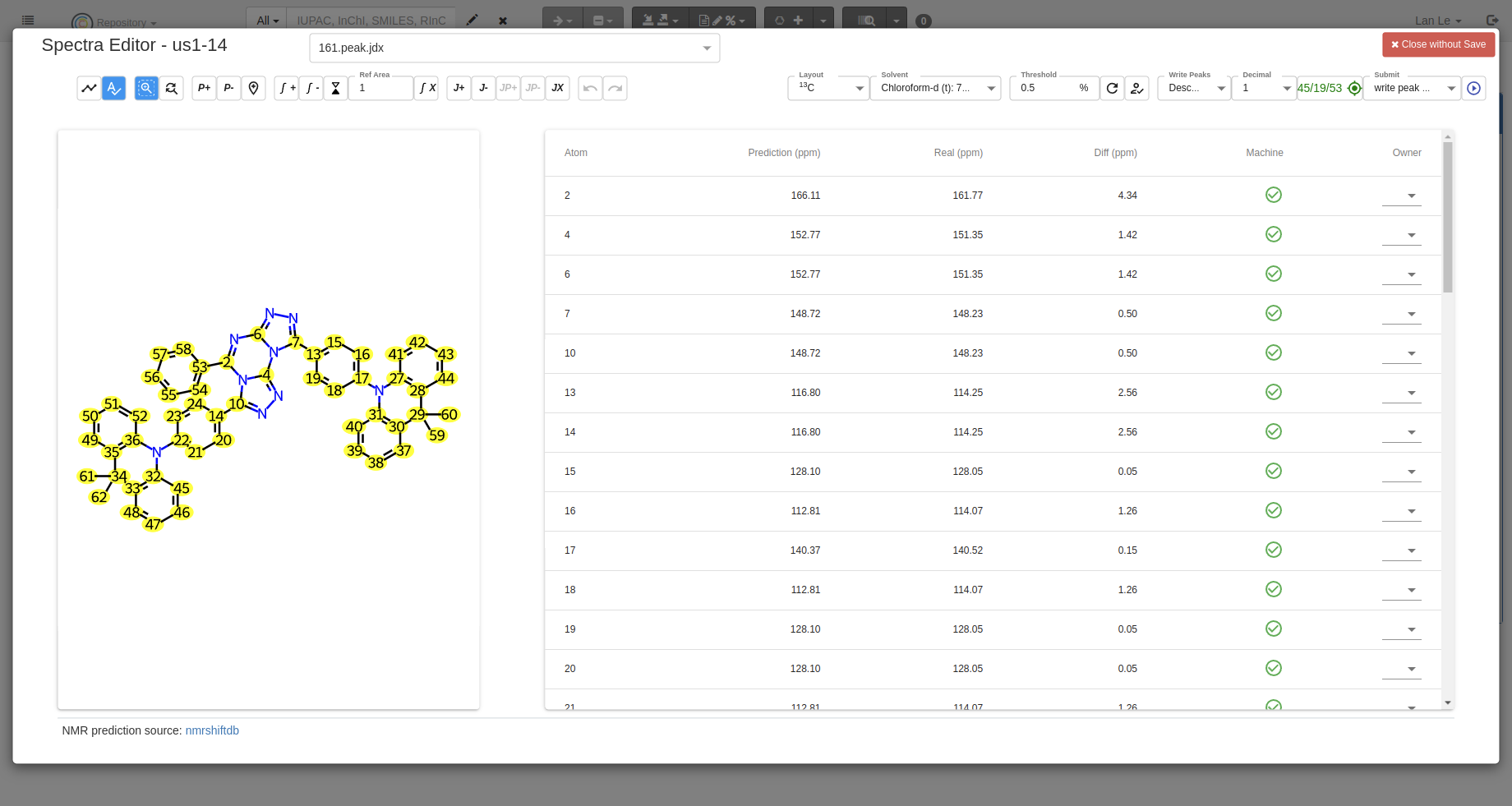
Predict NMR signals You can predict NMR signals with the
Predictbutton, the data of prediction comes from NMRshiftDB:
Analysis using NMRium
The official of the document of NMRium is located at https://docs.nmrium.org/
NMRium analysis files in Chemotion ELN are structured to optimise performance and loading time.
Analysis files do not embed the full spectral data. .nmrium files only contain references to the spectra and the analysis modifications applied by the user.
The raw spectral data is stored separately in .jdx files or .zip archives.
This separation results in smaller .nmrium files, faster analysis loading and reduced memory and network usage.
Demo Video
A video demonstrating how to use NMRium in Chemotion ELN
How-To
-
You can only open NMRium after defining that your
Type (Chemical Methods Ontology)is NMR: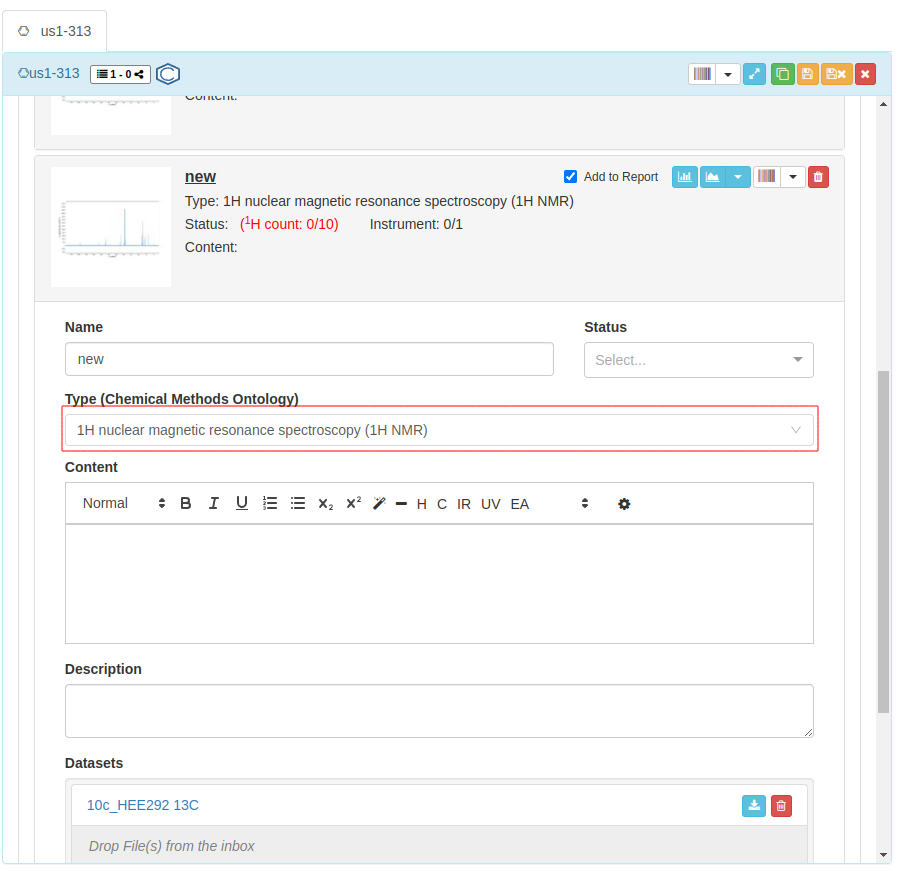
-
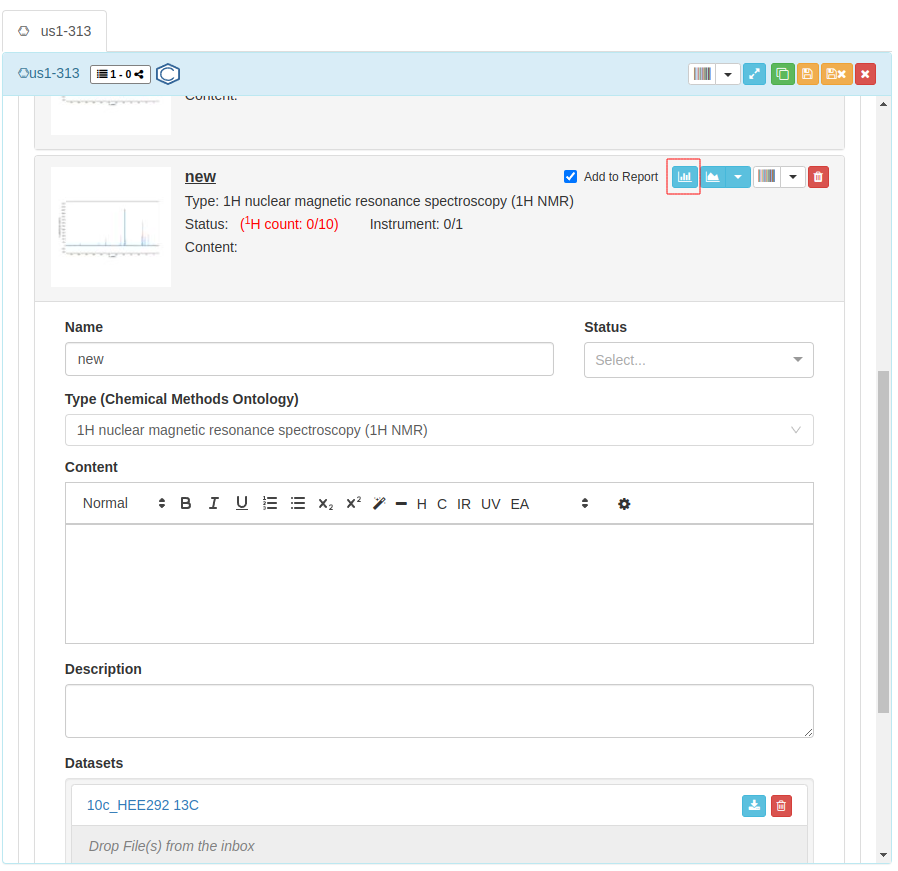
Click on the icon to open your file with NMRium.
-
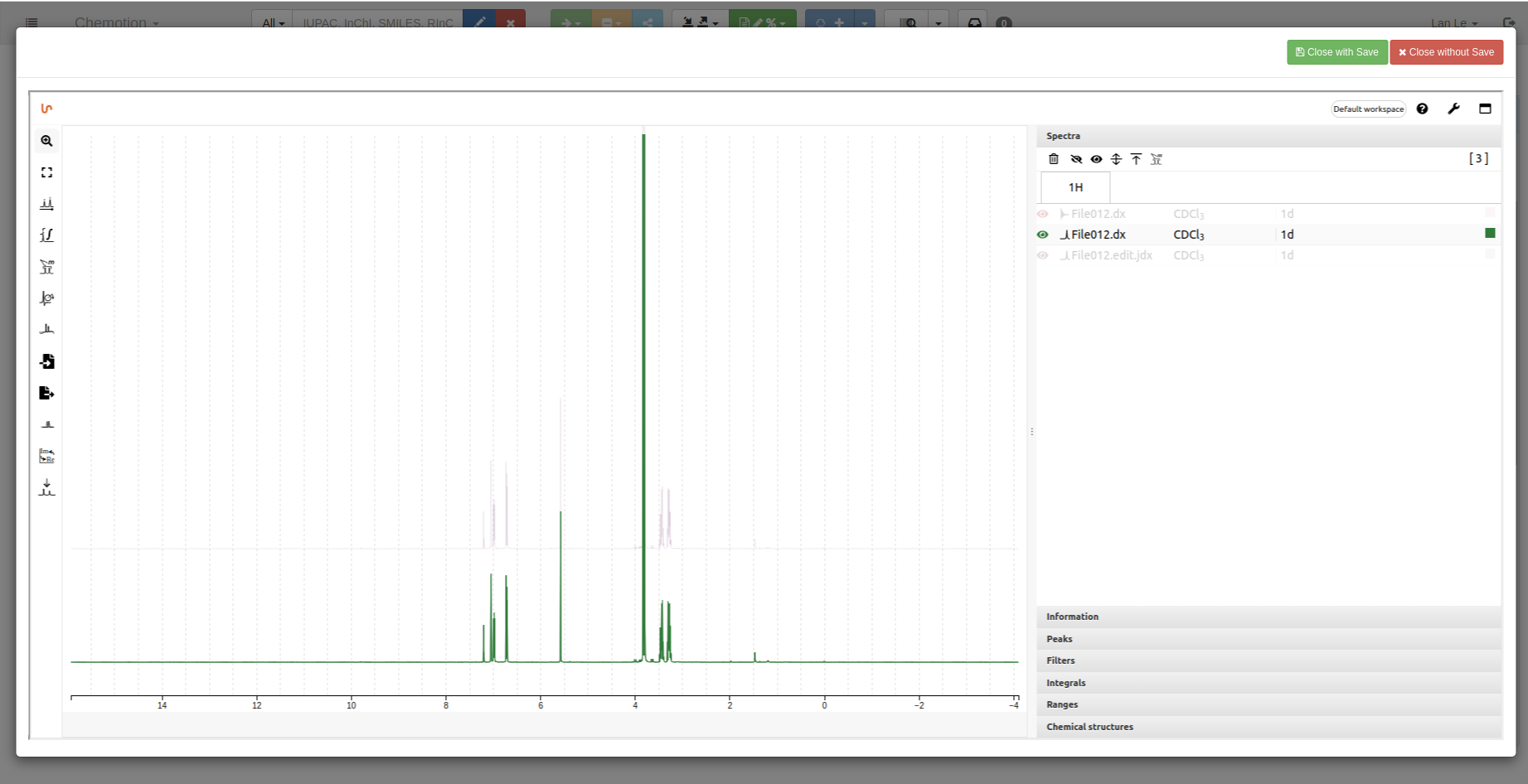
1D spectrum:
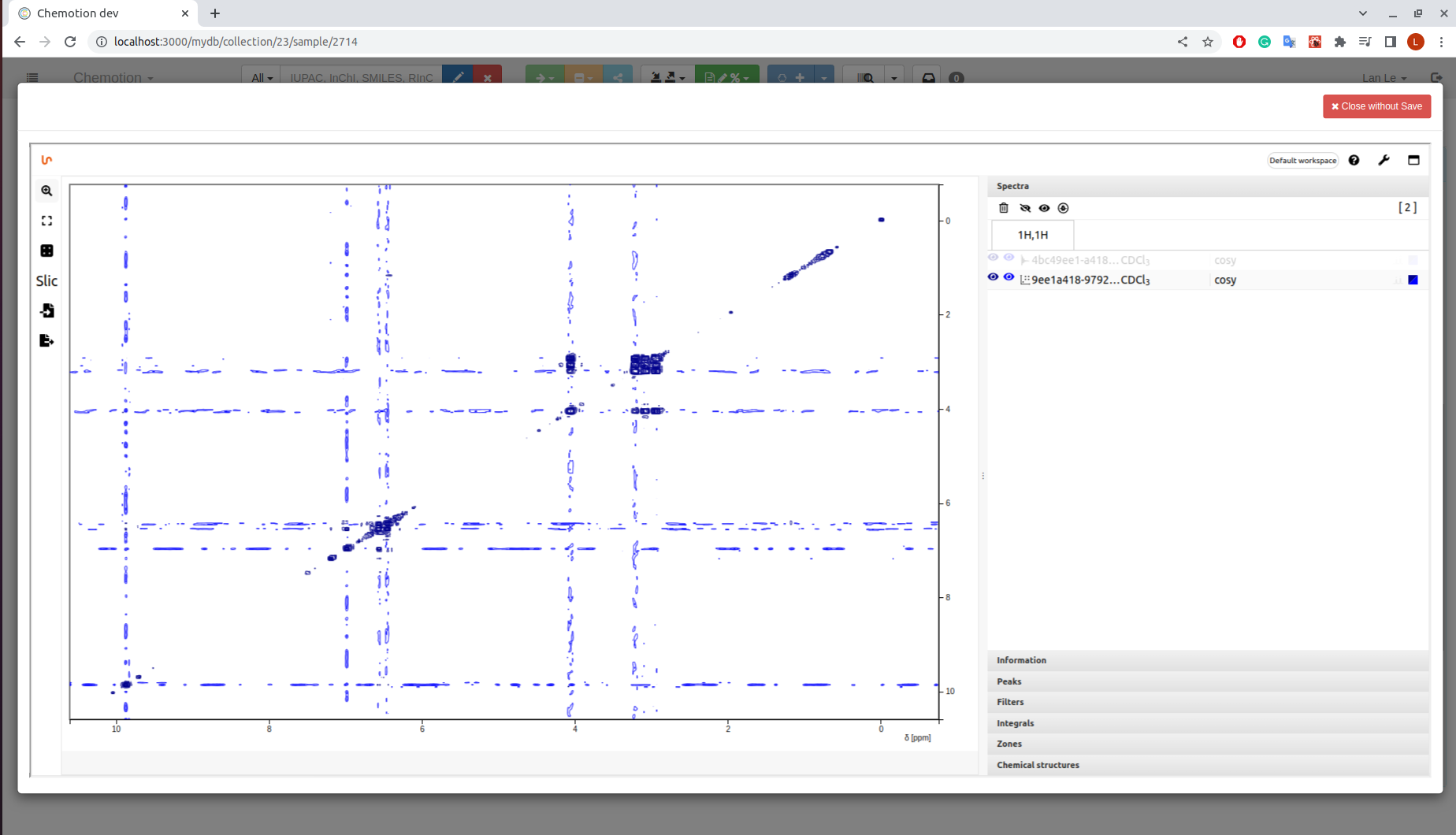
 2D spectrum
2D spectrum
-
Save your analyzed data by using
Close with Savebutton: